About Web-Resource
WheatQTLdb V2.0 is a manually curated QTL database for wheat that includes information about QTL identified through interval mapping and MTA identified using GWAS. The available information on metaQTL, epistatic QTL and candidate genes, wherever available, is also included in the database. Users could browse and download the database to find information regarding the genetic architecture of the traits of interest, which include the following: (i) tolerance to abiotic stresses including drought/water logging/heat/pre-harvest sprouting/salinity; (ii) resistance to biotic stresses including not only viral, bacterial and fungal diseases, but also to infestation by nematodes and insects; (iii) traits for biofortification (Fe/Se/Zn contents); (iv) developmental traits; (v) morphological traits; (vi) N/P/K use efficiency traits; (vii) physiological traits; (viii) quality traits; and (ix) yield and its related traits. WheatQTLdb is the largest web resource having collection of QTL (27,518), metaQTL (1,321), epistatic QTL (202) in Triticum aestivum and 07 other related wheat species to serve the international wheat research community including plant breeders and geneticists for further studies involving fine mapping, cloning and marker assisted selection (MAS) during wheat breeding.
How to cite us
If you use information gathered from this site in any publication, please cite the following paper:
Singh, K., Saini, D.K., Saripalli, G. et al. WheatQTLdb V2.0: a supplement to the database for wheat QTL. Mol Breeding 42, 56 (2022). https://doi.org/10.21203/rs.3.rs-1379009/v1
Singh K, Batra R, Sharma S, Saripalli G, Gautam T, Rakhi, Kumar J, Pal S, Malik P, Kumar M, Lateef I, Singh S, Kumar D, Saksham, Chaturvedi D, Verma A, Rani A, Kumar A, Sharma H, Jyoti, Kumar K, Kumar S, Singh VK, Singh VP, Kumar S, Kumar R, Sharma S, Gaurav SS, Sharma PK, Balyan HS, Gupta PK, 2020. WheatQTLdb: a QTL database for wheat. Mol Genet Genomics 296, 1051–1056 (2021). https://doi.org/10.1007/s00438-021-01796-9
Singh K, Batra R, Sharma S et al. (2021b) Correction to: WheatQTLdb: a QTL database for wheat. Mol Genet Genomics 296, 1359–1360. https://doi.org/10.1007/s00438-021-01812-y
Tutorial
|
|
Statistics
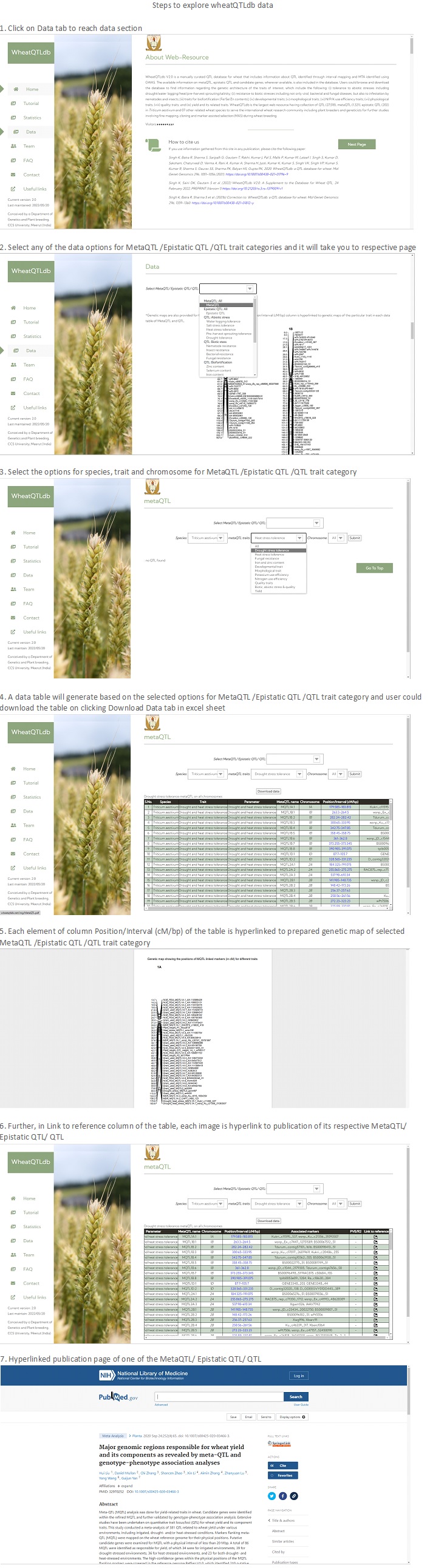
Frequencies of QTL, metaQTL and epistatic QTL in WheatQTLdb
Species wise distribution of QTL, metaQTL and epistatic QTL in wheatQTLdb
Overall QTL associated with various traits in WheatQTLdb
Chromosome wise distribution of QTL, metaQTL, epistatic QTL in Triticum aestivum
Year of publication wise frequencies of QTL, meta QTL and epistatic QTL
Data
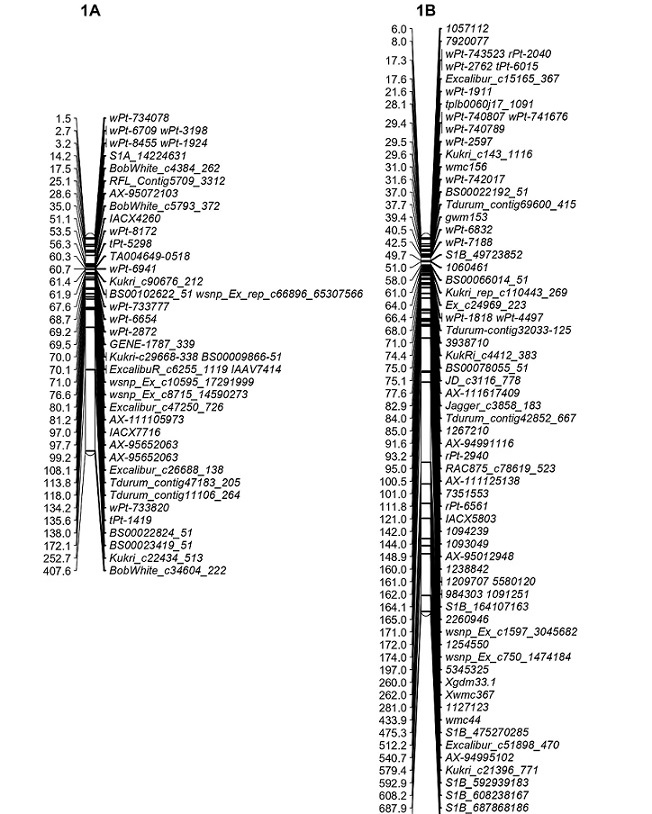
*Genetic maps are also provided for QTL and metaQTL.Each element in position/interval (cM/bp) column is hyperlinked to genetic maps of the particular trait in each data table of MetaQTL and QTL.
Team

Kalpana Singh MTech(Bioinfo), JRF, NET, GATE, PhD
Research Associate
Ch. Charan Singh University, Meerut (kalpana.iiita@gmail.com)

Dinesh Kumar Saini MSc, NET, GATE, ICAR-SRF
Research Scholar
Punjab Agricultural University, Ludhiana (dineshsaini96344@gmail.com)

Ritu Batra MSc(BioTech), NET, PhD
DS Kothari PostDoc Fellow
Ch. Charan Singh University, Meerut (ritu.biotech24@gmail.com)

Shiveta Sharma MSc(GPB), PhD
BioCARe Women Scientist
Ch. Charan Singh University, Meerut (s2sbhu@gmail.com)

Gautam Saripalli MSc(Ag), NET
Senior Research Fellow
Ch. Charan Singh University, Meerut (saripalligautam86@gmail.com)

Tinku Gautam MSc(BioTech), NET
Senior Research Fellow
Ch. Charan Singh University, Meerut (tinkugoutam@gmail.com)

Rakhi MSc(BioTech), MPhil, NET
Junior Research Fellow
Ch. Charan Singh University, Meerut (rakhi.7272@gmail.com)

Jitendra Kumar MSc(BioTech), SRF, NET, GATE, PhD
Research Associate
NABI, Mohali (jitendra_biotech09@rediffmail.com)

Sunita Pal MSc(BioTech), MPhil
Research Scholar
Ch. Charan Singh University, Meerut (dspal23@gmail.com)

Parveen Malik MSc(Ag), MPhil
Research Scholar
Ch. Charan Singh University, Meerut (parveenmalikccsu@gmail.com)

Manoj Kumar MSc(Ag), NET
Senior Research Fellow
Ch. Charan Singh University, Meerut (ch.manojmalik@gmail.com)

Irfat Jan MSc(BioTech), MPhil, NET
Junior Research Fellow
Ch. Charan Singh University, Meerut (irfatlateef1000@gmail.com)

Sahadev Singh MSc(Ag), MPhil
DST Inspire Fellow
Ch. Charan Singh University, Meerut (mr.sahadevsingh@gmail.com)

Deepti Chaturvedi MSc(BioTech)
Research Scholar
Ch. Charan Singh University, Meerut (deeptichaturvedi5@gmail.com)

Hemant Sharma MSc(Ag), MPhil
Junior Research Fellow
Ch. Charan Singh University, Meerut (sharmahemant150893@gmail.com)

Sourabh Kumar MSc(Ag), MPhil
Junior Research Fellow
Ch. Charan Singh University, Meerut (sourabhkumar7669@gmail.com)

Vivudh Pratap Singh MSc(Ag), MPhil
Junior Research Fellow
Ch. Charan Singh University, Meerut (vivudhpratap@gmail.com)

Deepak Kumar MSc(Botany), MPhil, NET, GATE
Research Scholar
Ch. Charan Singh University, Meerut (deepakpremi1991@gmail.com)

Saksham MSc(Botany), MPhil
Research Scholar
Ch. Charan Singh University, Meerut (www.sakshampundir@gmail.com)

Anuj Kumar MSc(Ag), MPhil, NET
Junior Research Fellow
Ch. Charan Singh University, Meerut (anuj2011.ag@gmail.com)

Kuldeep Kumar MSc(Ag), MPhil, NET
Junior Research Fellow
Ch. Charan Singh University, Meerut (kuldeepverma12395@gmail.com)

Anshu Rani MSc(BioTech), MPhil
Research Scholar
Ch. Charan Singh University, Meerut (anshu.bhattal@gmail.com)

Vikas Kumar Singh MSc(Ag)
DST Inspire Fellow
Ch. Charan Singh University, Meerut (vksgpb@gmail.com)

Anjali Verma MSc(BioTech), MPhil
Research Scholar
Ch. Charan Singh University, Meerut (anjalivr10795@gmail.com)
Jyoti MSc(BioTech)
Research Scholar
Ch. Charan Singh University, Meerut (jyotiku309@gmail.com)

Sachin Kumar MSc(Ag), PhD
Assistant Professor
Ch. Charan Singh University, Meerut (sachinkpsingh@gmail.com)

Rahul Kumar MSc(Ag),NET,MPhil, PhD
Associate Professor
Ch. Charan Singh University, Meerut (rahuldehran007@gmail.com)

Shailendra Sharma MSc(BioTech), NET, GATE, PhD
Professor & Head
Ch. Charan Singh University, Meerut (shgjus6@gmail.com)

Shailendra S Gaurav MSc(Ag), NET, PhD
Professor
Ch. Charan Singh University, Meerut (shgjus6@gmail.com)

Pradeep Kumar Sharma MSc(Ag), MPhil, NET, PhD
Professor
Ch. Charan Singh University, Meerut (pks264@rediffmail.com)

Harindra Singh Balyan PhD, FNASc, FNAAS, FNA
Hony. Emeritus Professor & INSA Senior Scientist
Ch. Charan Singh University, Meerut (hsbalyan@gmail.com)

PK Gupta PhD, FNASc, FNAAS, FASc, FNA
Hony. Emeritus Professor & INSA Senior Scientist
Ch. Charan Singh University, Meerut (pkgupta36@gmail.com)
FAQ
1. What are QTL/MTA?
Quantitative Trait Loci (QTL) are specific chromosomal regions containing genes that make a significant additive contribution to the expression of a complex trait; identified by, analyzing or comparing the linkage (degree of co-variation) of polymorphic molecular markers and variation of phenotypic traits. Marker trait association (MTA) is whole genome scan using GWAS of association between genetic markers (eg. SNP) and phenotypes with specific family structure designed for such analysis with certain traits. Therefore, both QTL and MTA are genomic mappings of traits. In WheatQTLdb, we treated both as QTL.
2. What is WheatQTLdb? Is there any publication about it?
WheatQTLdb is an abbreviated name for wheat QTL database, which contains published QTL/MTA curated into structured tables managed in a relational database environment, MySQL. The user interface with web-server is programmed with PHP. The progress on the WheatQTLdb development has been presented at National Symposium on Database Development and Biocuration organized by Department of Plant Molecular Biology, University of Delhi South Campus, New Delhi, India (December 17-18, 2019). Later, published in Mol Genet Genomics (https://doi.org/10.1007/s00438-021-01796-9) & (https://doi.org/10.1007/s00438-021-01812-y).
3. Which species are included in WheatQTLdb?
In WheatQTLdb V1.0 only hexaploid wheat (Triticum aestivum) has been included, later we extended it in V2.0 to include the information related to 07 other diploid/tetraploid wheat species (T. durum, T. monococcum, T. boeoticum, T. turgidum, T. dicoccoides, T. dicoccum and Aegilops tauschii) as well.
4. How are traits classified in WheatQTLdb?
To serve the wheat research community in a better way, we grouped traits into functional categories to be directly used for a particular reserach topic as lolerance to water logging, salt stress, drought, pre-harvest sprouting etc., morphological, quality, yield, developmental, physiological, resistance to bacterial, fungal, nematode, insect diseases, N/P use efficiency.
5. How are public QTL data curated into WheatQTLdb?
Following information was extracted from published literature: Species, Trait, Parameter, Cross, Population/Germplasm(size), Method, QTL name, Chromosome, Position (interval) in cM/bp, Associated markers, PVE/R2, Candidate gene and reference.
6. Can I enter my QTL data into WheatQTLdb?
Yes. WheatQTLdb is open to public for data entry, anyone interested could send us data as attachment in .csv/.xlsx file format through the provided form on contact page or directly mail us.
Contact Us
In case you have any query or would like to add any data in our database, kindly contact us. We will please to respond or include the provided data in our database.
Our Location
View Larger Map
Mailing Address
Contact Person
Prof PK Gupta,Hony. Emeritus Professor & INSA Hony. Scientist,
Ch. Charan Singh University, Meerut (INDIA)
Phone: +919411619105
Email: pkgupta36@gmail.com